Equivariant Diffusion for Molecule Generation in 3D using Consistency Models
Introduction to the seminal papers "Equivariant Diffusion for Molecule Generation in 3D" and "Consistency Models" with an adaptation fusing the two together for fast molecule generation.
Introduction
In this blog post, we discuss the paper “Equivariant Diffusion for Molecule Generation in 3D”
Most diffusion models are unfortunately bottle-necked by the sequential denoising process, which can be slow and computationally expensive
Briefly on Equivariance for molecules
Equivariance is a property of certain functions, which ensures that their output transforms in a predictable manner under collections of transformations. This property is valuable in molecular modeling, where it can be used to ensure that the properties of molecular structures are consistent with their symmetries in the real world. Specifically, we are interested in ensuring that structure is preserved in the representation of a molecule under three types of transformations: translation, rotation, and reflection.
Formally, we say that a function \(f\) is equivariant to the action of a group \(G\) if:
\[\begin{align} T_g(f(x)) = f(S_g(x)) \end{align}\]for all \(g \in G\), where \(S_g,T_g\) are linear representations related to the group element \(g\)
The three transformations: translation, rotation, and reflection, form the Euclidean group \(E(3)\), which is the group of all aforementioned isometries in three-dimensional space, for which \(S_g\) and \(T_g\) can be represented by a translation \(t\) and an orthogonal matrix $R$ that rotates or reflects coordinates.
A function \(f\) is then equivariant to a rotation or reflection \(R\) if transforming its input results in an equivalent transformation of its output
Introducing Equivariant Graph Neural Networks (EGNNs)
Molecules can very naturally be represented with graph structures, where the nodes are atoms and edges their bonds. The features of each atom, such as its element type or charge can be encoded into an embedding \(\mathbf{h}_i \in \mathbb{R}^d\) alongside with the atoms 3D position \(\mathbf{x}_i \in \mathbb{R}^3\).
To learn and operate on such structured inputs, Graph Neural Networks (GNNs)

The previously mentioned \(E(3)\) equivariance property of molecules can be injected as an inductive prior into to the model architecture of a message passing graph neural network, resulting in an \(E(3)\) EGNN. This property improves generalisation
The EGNN is built with equivariant graph convolution layers (EGCLs):
\[\begin{align} \mathbf{x}^{l+1},\mathbf{h}^{l+1}=EGCL[ \mathbf{x}^l, \mathbf{h}^l ] \end{align}\]An EGCL layer can be formally defined by:
where \(h_l\) represents the feature $h$ at layer \(l\), \(x_l\) represents the coordinate at layer \(l\) and \(d_{ij}= ||x_i^l-x^l_j||_2\) is the Euclidean distance between nodes \(v_i\) and \(v_j\).
A fully connected neural network is used to learn the functions \(\phi_e\), \(\phi_x\), and \(\phi_h\). At each layer, a message \(m_{ij}\) is computed from the previous layer’s feature representation. Using the previous feature and the sum of these messages, the model computes the next layer’s feature representation.
This architecture then satisfies translation and rotation equivariance. Notably, the messages depend on the distance between the nodes and these distances are not changed by isometric transformations.
Equivariant Diffusion Models (EDM)
This section introduces diffusion models and describes how their predictions can be made \(E(3)\) equivariant. The categorical properties of atoms are already invariant to \(E(3)\) transformations, hence, we are only interested in enforcing this property on the sampled atom positions.
What are Diffusion Models?
Diffusion models

The “forward” noising process can be parameterized by a Markov process
The whole Markov process leading to time step \(T\) is given as a chain of these transitions:
\[\begin{align} q\left( x_1, \ldots, x_T \mid x_0 \right) := \prod_{t=1}^T q \left( x_t \mid x_{t-1} \right) \end{align}\]The “reverse” process transitions are unknown and need to be approximated using a neural network parametrized by \(\theta\):
\[\begin{align} p_\theta \left( x_{t-1} \mid x_t \right) := \mathcal{N} \left( x_{t-1} ; \mu_\theta \left( x_t, t \right), \Sigma_\theta \left( x_t, t \right) \right) \end{align}\]Because we know the dynamics of the forward process, the variance \(\Sigma_\theta \left( x_t, t \right)\) at time \(t\) is known and can be fixed to \(\beta_t \mathbf{I}\).
The predictions then only need to obtain the mean \(\mu_\theta \left( x_t, t \right)\), given by:
\[\begin{align} \mu_\theta \left( x_t, t \right) = \frac{1}{\sqrt{\alpha_t}} \left( x_t - \frac{\beta\_t}{\sqrt{1 - \bar{\alpha}\_t}} \epsilon\_\theta \left( x_t, t \right) \right) \end{align}\]where \(\alpha_t = \Pi_{s=1}^t \left( 1 - \beta_s \right)\).
Hence, we can directly predict \(x_{t-1}\) from \(x_{t}\) using the network \(\theta\):
\[\begin{align} x_{t-1} = \frac{1}{\sqrt{1 - \beta_t}} \left( x_t - \frac{\beta_t}{\sqrt{1 - \alpha_t}} \epsilon_\theta \left( x_t, t \right) \right) + \sqrt{\beta_t} v_t \end{align}\]where \(v_T \sim \mathcal{N}(0, \mathbf{I})\) is a sample from the pure Gaussian noise.
Enforcing E(3) equivariant diffusion
Equivariance to rotations and reflections effectively means that if any orthogonal rotation matrix \(\mathbf{R}\) is applied to a sample \(\mathbf{x}_t\) at any given time step \(t\), we should still generate a correspondingly rotated “next best sample” \(\mathbf{R}\mathbf{x}_{t+1}\) at time \(t+1\).
In other words, the likelihood of this next best sample does not depend on the molecule’s rotation and the probability distribution for each transition in the Markov Chain is hence roto-invariant:
\[\begin{align} p(y|x) = p(\mathbf{R}y|\mathbf{R}x) \end{align}\]
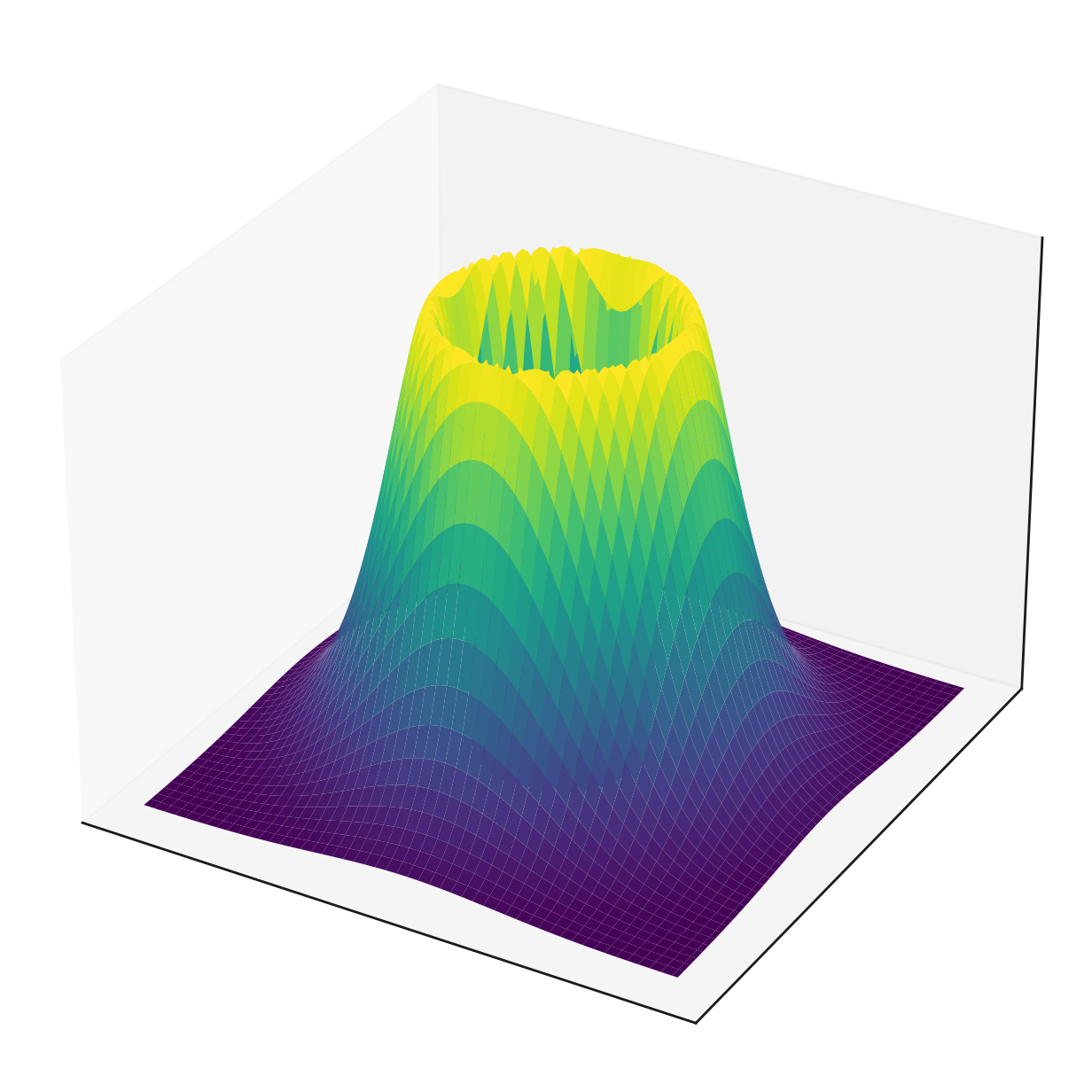
Figure 3: Examples of 2D roto-invariant distributions
Such an invariant distribution composed with an equivariant invertible function results in another invariant distribution
Since the underlying EGNN already ensures this equivariance, the remaining constraint can easily be achieved by setting the initial sampling distribution to something roto-invariant, such as a simple mean zero Gaussian with a diagonal covariance matrix, as illustrated in Figure 3 (left).
Translation equivariance requires a few tricks. It has been shown, that it is impossible to have non-zero distributions invariant to translations
The EDM authors bypass this with a clever trick of always re-centering the generated samples to have center of gravity at \(\mathbf{0}\) and further show that these \(\mathbf{0}\)-centered distributions lie on a linear subspace that can reliably be used for equivariant diffusion
How to train the EDM?
The training objective of diffusion-based generative models amounts to “maximizing the log-likelihood of the sample on the original data distribution.”
During training, a diffusion model learns to approximate the parameters of a posterior distributions at the next time step by minimizing the KL divergence between this estimate and the ground truth, which is equivalent objective to minimizing the negative log likelihood.
\[\begin{align} L_{vlb} := L_{t-1} := D_{KL}(q(x_{t-1}|x_{t}, x_{0}) \parallel p_{\theta}(x_{t-1}|x_{t})) \end{align}\]The EDM adds a caveat that the predicted distributions must be calibrated to have center of gravity at \(\mathbf{0}\), in order to ensure equivariance.
Using the KL divergence loss term with the EDM model parametrization simplifies the loss function to:
\[\begin{align} \mathcal{L}_t = \mathbb{E}_{\epsilon_t \sim \mathcal{N}_{x_h}(0, \mathbf{I})} \left[ \frac{1}{2} w(t) \| \epsilon_t - \hat{\epsilon}_t \|^2 \right] \end{align}\]where \(w(t) = \left(1 - \frac{\text{SNR}(t-1)}{\text{SNR}(t)}\right)\) and \(\hat{\epsilon}_t = \phi(z_t, t)\).
The EDM authors found that the model performs best with a constant \(w(t) = 1\), thus effectively simplifying the loss function to an MSE. Since coordinates and categorical features are on different scales, it was also found that scaling the inputs before inference and then rescaling them back also improves performance.
Consistency Models
As previously mentioned, diffusion models are bottlenecked by the sequential denoising process
Modelling the noising process as an SDE
Song et al.
Where \(t\) is the time-step, \(\mathbf{\mu}\) is the drift coefficient, \(\sigma\) is the diffusion coefficient, and \(\mathbf{w}_t\) is the stochastic component denoting standard Brownian motion. This stochastic component effectively represents the iterative adding of noise to the data in the forward diffusion process and dictates the shape of the final distribution at time \(T\).
Typically, this SDE is designed such that \(p_T(\mathbf{x})\) at the final time-step \(T\) is close to a tractable Gaussian.

Existence of the PF ODE
This SDE has a remarkable property, that a special ODE exists, whose trajectories sampled at \(t\) are distributed according to \(p_t(\mathbf{x})\)
This ODE is dubbed the Probability Flow (PF) ODE by Song et al.
A score model \(s_\phi(\mathbf{x}, t)\) can be trained to approximate \(\nabla log p_t(\mathbf{x})\) via score matching
With \(\mathbf{\hat{x}}_T\) sampled from the specified Gaussian at time \(T\), the PF ODE can be solved backwards in time to obtain a solution trajectory mapping all points along the way to the initial data distribution at time \(\epsilon\) very close to zero.
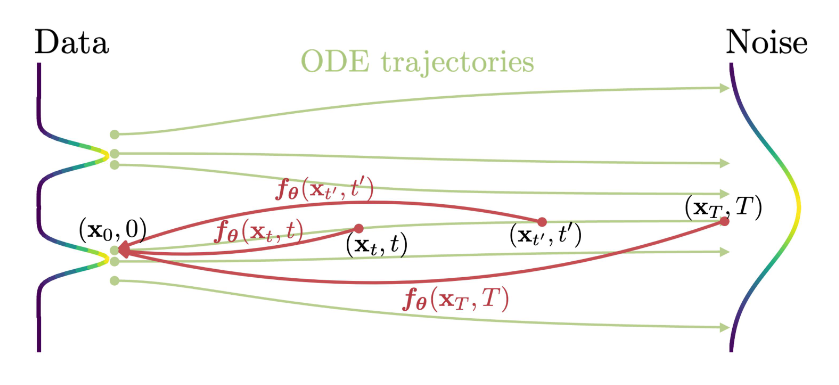
Given any off-the-shelf ODE solver (e.g. Euler) and a trained score model \(s_\phi(\mathbf{x}, t)\), we can solve this PF ODE. The time horizon \([\epsilon, T]\) is discretized into sub-intervals for improved performance
Consistency Function
Given a solution trajectory \({\mathbf{x}_t}\), we define the consistency function as:
\[\begin{align} f: (\mathbf{x}_t, t) \to \mathbf{x}_{\epsilon} \end{align}\]In other words, for every pair (\(\mathbf{x}_t\), \(t\)), a consistency function always outputs a corresponding datapoint at time $\epsilon$, which will be very close to the original data distribution.
Importantly, this function has the property of self-consistency: i.e. its outputs are consistent for arbitrary pairs of \((x_t, t)\) that lie on the same PF ODE trajectory. Hence, we have
\[f(x_t, t) = f(x_{t'}, t') \text{ for all } t, t' \in [\epsilon, T]\]The goal of a consistency model, denoted by \(f_\theta\), is to estimate this consistency function \(f\) from data by being enforced with this self-consistency property during training.
Sampling
With a fully trained consistency model \(f_\theta(\cdot, \cdot)\), we can generate new samples by simply sampling from the initial Gaussian \(\hat{x_T}\) \(\sim \mathcal{N}(0, T^2I)\) and propagating this through the consistency model to obtain samples on the data distribution \(\hat{x_{\epsilon}}\) \(= f_\theta(\hat{x_T}, T)\) with as little as one diffusion step.

Training Consistency Models
Consistency models can either be trained by “distillation” from a pre-trained diffusion model, or in “isolation” as a standalone generative model from scratch. In the context of our work, we focused only on the latter because the distillation approach has a hard requirement of using a pretrained score based diffusion. In order to train in isolation we need to leverage the following unbiased estimator:
\[\begin{align} \nabla \log p_t(x_t) = - \mathbb{E} \left[ \frac{x_t - x}{t^2} \middle| x_t \right] \end{align}\]where \(x \sim p_\text{data}\) and \(x_t \sim \mathcal{N}(x; t^2 I)\).
That is, given \(x\) and \(x_t\), we can estimate \(\nabla \log p_t(x_t)\) with \(-(x_t - x) / t^2\). This unbiased estimate suffices to replace the pre-trained diffusion model in consistency distillation when using the Euler ODE solver in the limit of \(N \to \infty\)
Song et al.
where \(\mathbf{z} \sim \mathcal{N}(0, I)\).
Crucially, \(\mathcal{L}(\theta, \theta^-)\) only depends on the online network \(f_\theta\), and the target network \(f_{\theta^-}\), while being completely agnostic to diffusion model parameters \(\phi\).
Experiments
We replicate the original EDM set-up and evaluate on the QM9 dataset
| Model / Sampling Time (seconds) | Mean | STD |
|---|---|---|
| Default EDM | 0.6160 | 0.11500 |
| Consistency Model (single step) | 0.0252 | 0.00488 |
As expected, we observed in table 1., that the consistency model in single-step mode is significantly faster than the EDM, providing up to a 24x speed-up averaged over 5 sampling runs. This number represents the time it takes the model to generate a sample on the data distribution from a pure Gaussian noise input, excluding other computational overheads shared by both models equally, such as logging.
| Model / Metric | Training NLL | Validation NLL | Best Cross-Validated Test NLL | Best Atom Stability | Best Molecule Stability |
|---|---|---|---|---|---|
| Default EDM | 2.524 | -30.066 | -17.178 | 0.873 | 0.196 |
| Consistency Model (single step) | 2.482 | 94176 | 80363 | 0.19 | 0 |
| Consistency Model (multi-step) | 2.484 | 166264 | 179003 | 0.12 | 0 |
We observed that the consistency models converge on the training set with similar rate as the regular EDM, even achieving slightly lower training NLLs. However, they completely fail to generalize to the validation and test sets with much lower atom stability (the proportion of atoms that have the right valency) and no molecule stability (the proportion of generated molecules for which all atoms are stable). These results are surprisingly poor, given that the dataset is not particularly complicated, and consistency models have already shown promising results on images
To improve these results, we attempted to use multi-step sampling, which should in theory allow us to replicate results close to the EDM with the same number of sampling steps. However, we observed no such improvement in our experiments. We tested multiple different amounts of steps and report results for 100, which performed best overall. Oddly, the multi-step sampling actually yields worse results than the single-step sampling most of the time, which is highly unexpected and requires further investigation.
It should also be noted that the default EDM with more training is capable of achieving results much better than what we report in table 2. However, it still comfortably outperforms all consistency model variations on all metrics using equal amounts of compute.
Discussion
Consistency models are able to reduce the number of steps during sampling up to just a single step, significantly speeding up the sampling process. We were able to successfully demonstrate this and train an EDM as a consistency model in isolation, achieving nearly identical training loss with up to 24x faster sampling times. However, using the single-step sampling only achieves up to 19% atom stability in best case scenario, compared with the default EDM which consistently reaches 87% or much more with further training. We suspect that a model trained in this set-up might be too prone to overfitting and struggles with generalization to anything outside the training data distribution, compared to sequential de-noising predictions of the EDM, which are more robust by design.
Using multi-step sampling should in theory yield competitive results, but we observed no such improvement. Since it cannot be conclusively ruled out that this was caused by a bug in our multi-step sampling code, we hope to continue investigating if the consistency model paradigm can reliably be used for molecule generation in the future and show more competitive results as previous works suggest is possible